By Jonas Waldenström
I just finished a course in epidemics at Penn State University. But without setting my foot on campus, or, in fact USA! And guess what, I didn’t miss a single lecture! Sounds weird? Thing is, this was not an ordinary course; this was a MOOC – the latest development in distance learning. And it was great! But more on that later.
We live in an ongoing digital and technical revolution – just compare everyday life today with how it was ten, or twenty years ago. The change is not linear; rather it is a series of punctuated shifts, where each new wonder gadget push the field to a new plateau. For instance, back in 1999 when I worked in northern Nigeria it was virtually impossible to phone home. In the bigger cities you could pay for a call via landlines, but in the area where we worked the closest phone was 10 hours away, across the sand dunes. A few years later mobile phones were everywhere, and Internet cafes were popping up on the posh streets. In fact, these days, you often have better connection between Nigeria and Sweden, than between Kalmar and Öland…

The hallmark of an obsolete technique
Another example, although more linear, is the ever-faster computer processors, or the speed at which you can sequence DNA. When I started my PhD, the art of drawing figures by hand, in ink, for publications was still remembered. And during my first year I was extremely happy if I managed to run 10 sequences (of roughly 500 bp each) over night on the capillary sequencer! Now you can print a gun in 3D, or sequence a genome over night. At least a microbial genome, and a very simple gun. But still, who would have believed this 20 year ago?
Education, however, has evolved at a slower pace, even after the onset of computers. Yes, some guy invented PowerPoint in 1987 and sold it to Microsoft, thereby turning a bad lecture into a worse, but more colorful lecture. (Actually, the ‘internets’ tell me they were two guys: Robert Gaskins and Dennis Austin, and that they both think their creation has sometimes turned into an abomination.) Otherwise the key concepts of a lecture have remained the same for many centuries. Teaching is primarily occurring here and now, in a room of varying size at a campus. Teacher goes in, does his/her thing and hopefully the students become enthusiastic, interact and learn. A familiar process for all of us, unless you drop out of school at the age of seven. A good lecture can change your purpose of life. On the other hand, a bad lecture can be akin to torture and be remembered for far too long.
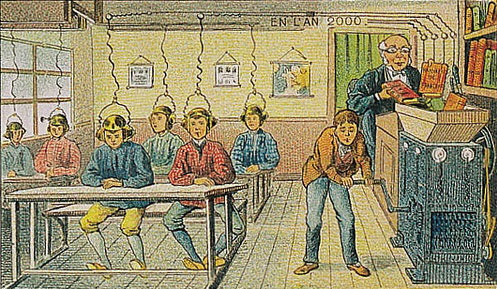
The future of education as seen in 1905
The question now is whether this is about to change? At least, for the first time it seems as technology is there, or nearly there, to allow things to be done differently. And this is where the MOOC – the Massive Online Open Courses – comes in to play. Education for the masses, free and online. Some believe this way of teaching will make education democratic and open it for everyone, not only the fortunate who by smartness or rich parents study at the prestigious universities. Via a connection to the Internet it is now possible to enroll in courses on a vast number of subjects. A smorgasbord of education. Through sites as Coursera, the disparate courses can be collected into a degree; tailored and individual-based degrees.
So what about the epidemics MOOC? Truly, it was a magnificent course! The team of teachers was like a NHL team of only seriously talented scientists: Dr. Marcel Salathé, Dr. Ottar N. Bjornstad, Dr. Rachel A. Smith, Dr. Mary L. Poss, Dr. David P. Hughes, Dr. Peter Hudson, Dr. Matthew Ferrari, and Dr. Andrew Read. And it was indeed a massive course, even for a MOOC! One figure that was mentioned was 27,000 enrolled students! One year I gave my course, with a rather similar content, to 6 students – that tells you of the potential outreach a well-managed online course can have compared to a campus course at a small university.

A screen shot of Dr Andrew Read in action.
A MOOC, or any type of distance learning, is as good as the teachers are, and the time and resources put in by the university. The Epidemics MOOC was certainly an extremely good example, where it was evident that they wanted to do something more than the ordinary, set a new standard even. However, even if the course was beautifully made and stuffed with interesting lectures, I reckon that there must have been a massive drop out of students. Not because the course was too advanced, but because it is so easy to procrastinate things to a point where you have several weeks of lectures to digest. A human fallacy, I’d say. Also, I guess that a significant fractions of students, and perhaps over-represented among those that finished it, were folks like me. Folks that already have a degree, or even work with the subject. But that doesn’t matter too much after all – if only 10% of the students made it all the way, it still means that a staggering 2,700 people now are acquainted with the basics of epidemiology!
In case Marcel Salathé (or any of the other teachers) reads this little piece, I’d like to end with some reflections of the course:
You understood your audience. Most people that are enrolled in distance education are already busy. If you have a day job you don’t have all the time in the world. You want things to be condensed, edited and thoroughly thought through. In my case I watched the videos when my youngest daughter was napping – which isn’t forever, I can tell you. Instead of long lectures there were several short (5 – 9 min each); short enough to maintain focus, and long enough to say something important. Well done!
We could see you! I love the fact that the videos were not uploaded powerpoints with voice-over. These were actual lectures recorded in studio, where the lecturer looked into the camera. Instead of the ppt, the main notes were highlighted via cartoons appearing together with the talker. That’s smart!
I could rewind, skip, press forward, and even adjust the speed of the talks. The possibility to watch lectures when you want to is great, of course. And that you can download them and watch when you are offline too. I also liked the speed button. The Andrew Read NZ dialect required a 0.75 speed setting, while Ottar Bjornstad and Mary Poss were best enjoyed at a 1.25 pace. Splendid idea!
Ask us anything. The idea of picking questions from the study forums and have group discussion among the faculty was an excellent idea. And a possibility to pick up recent events like flu and MERS. I liked this, good move.
The heterogeneity of students was a problem, at least for discussions. A central idea is that the platform should stimulate discussion via online forums. For this course it was expected that the student engaged in at least 10 posts to earn credits. However, with 27,000 students enrolled the level of comments was often more Facebookish than intellectual. Perhaps it would have been good to add a few explanatory lectures (like a week zero) describing the very basics (such as what a virus really is).
With that said, I really enjoyed the course! Whether the MOOCs will change education as we know it is still an open question though. We will see what the future brings! Or to paraphrase Reel 2 Real: MOOC it, MOOC it!
*******************************************************************************************************************
If you enjoyed this post, or other posts on this blog, why not follow the blog via email, Feedly or get updates via Twitter by following @DrSnygg?